Date: 20 Sep 2019 Title: Goodtables
Today, we’ll see how FrictionlessDarwinCore and Goodtables can help GBIF users with a simple data flow:
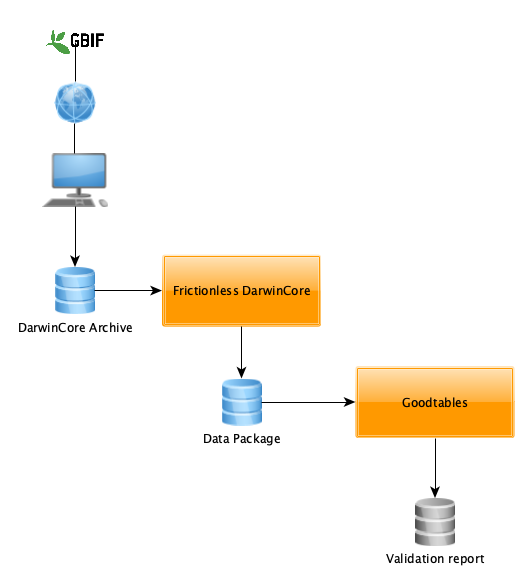
1. Download occurrences from GBIF portal
Login to GBIF.org Data portal.
Create and refine your query with the appropriate filters.
Download the resulting occurrences in DarwinCore format.
Store this DwC archive on your HD, eg under 0000637-190918142434337.zip
2. Convert this DwCA into a DataPackage
$ fdwca 0000637-190918142434337.zip download_DP.zip
...
3. Validate downloaded occurrences with goodtables
$ goodtables download_DP.zip
DATASET
=======
{'error-count': 8560,
'preset': 'nested',
'table-count': 3,
'time': 18.842,
'valid': False}
TABLE [1]
=========
{'datapackage': 'download_DP.zip',
'error-count': 8533,
'format': 'inline',
'headers': ['gbifID',
'abstract',
'accessRights',
...
'lastParsed',
'lastCrawled',
'repatriated'],
'row-count': 4416,
'schema': 'table-schema',
'source': '/var/folders/19/mrkkz1tx34l98n1rs3qr3xr00000gn/T/tmpb7ugs_ug-datapackage/occurrence.txt',
'time': 18.468,
'valid': False}
---------
[2,64] [enumerable-constraint] The value "PRESERVED_SPECIMEN" in row 2 and column 64 does not conform to the given enumeration: "['PreservedSpecimen', 'FossilSpecimen', 'LivingSpecimen', 'MaterialSample', 'Event', 'HumanObservation', 'MachineObservation', 'Taxon', 'Occurrence']"
[25,64] [enumerable-constraint] The value "HUMAN_OBSERVATION" in row 25 and column 64 does not conform to the given enumeration: "['PreservedSpecimen', 'FossilSpecimen', 'LivingSpecimen', 'MaterialSample', 'Event', 'HumanObservation', 'MachineObservation', 'Taxon', 'Occurrence']"
[25,135] [type-or-format-error] The value "3536.0" in row 25 and column 135 is not type "integer" and format "default"
...
TABLE [2]
=========
{'datapackage': 'download_DP.zip',
'error-count': 27,
'format': 'inline',
'headers': ['gbifID',
'abstract',
'accessRights',
...
'taxonomicStatus',
'nomenclaturalStatus',
'taxonRemarks'],
'row-count': 4416,
'schema': 'table-schema',
'source': '/var/folders/19/mrkkz1tx34l98n1rs3qr3xr00000gn/T/tmpb7ugs_ug-datapackage/verbatim.txt',
'time': 17.675,
'valid': False}
---------
[103,123] [maximum-length-constraint] The value "BEL" in row 103 and column 123 does not conform to the maximum length constraint of "2"
[470,36] [maximum-length-constraint] The value "Various" in row 470 and column 36 does not conform to the maximum length constraint of "2"
[470,142] [type-or-format-error] The value "707.1" in row 470 and column 142 is not type "integer" and format "default"
[1440,64] [enumerable-constraint] The value "ObservedSpecimen" in row 1440 and column 64 does not conform to the given enumeration: "['PreservedSpecimen', 'FossilSpecimen', 'LivingSpecimen', 'MaterialSample', 'Event', 'HumanObservation', 'MachineObservation', 'Taxon', 'Occurrence']"
[1441,64] [enumerable-constraint] The value "ObservedSpecimen" in row 1441 and column 64 does not conform to the given enumeration: "['PreservedSpecimen', 'FossilSpecimen', 'LivingSpecimen', 'MaterialSample', 'Event', 'HumanObservation', 'MachineObservation', 'Taxon', 'Occurrence']"
[1618,123] [maximum-length-constraint] The value "BE-WNA" in row 1618 and column 123 does not conform to the maximum length constraint of "2"
[4416,64] [enumerable-constraint] The value "Museum specimen" in row 4416 and column 64 does not conform to the given enumeration: "['PreservedSpecimen', 'FossilSpecimen', 'LivingSpecimen', 'MaterialSample', 'Event', 'HumanObservation', 'MachineObservation', 'Taxon', 'Occurrence']"
...
TABLE [3]
=========
{'datapackage': 'download_DP.zip',
'error-count': 0,
'format': 'inline',
'headers': ['gbifID',
'type',
'format',
'identifier',
'references',
'title',
'description',
'created',
'creator',
'contributor',
'publisher',
'audience',
'source',
'license',
'rightsHolder'],
'row-count': 176,
'schema': 'table-schema',
'source': '/var/folders/19/mrkkz1tx34l98n1rs3qr3xr00000gn/T/tmpb7ugs_ug-datapackage/multimedia.txt',
'time': 0.076,
'valid': True}
4. If necessary, add user constraints
Open data_package.json (from download_DP.zip), and add your own user constraints such as:
- individualCount (column 72) is required
- coordinateUncertaintyInMeters (column 135) maximum value is 1000
- coordinatePrecision (column 136) is required
See Tabular Data Specifications for more details on Table Schema, column type, format and constraints
"resources": [ { "name": "occurrence", "path": "occurrence.txt", "description": "http://rs.tdwg.org/dwc/terms/Occurrence", "schema": { "fields": [ { "name": "individualCount", "type": "integer", "description": "http://rs.tdwg.org/dwc/terms/individualCount", "constraints": { "required": true } }, { "name": "coordinateUncertaintyInMeters", "type": "number", "description": "http://rs.tdwg.org/dwc/terms/coordinateUncertaintyInMeters", "constraints": { "maximum": 1000.0 } }, { "name": "coordinatePrecision", "type": "number", "description": "http://rs.tdwg.org/dwc/terms/coordinatePrecision", "constraints": { "required": true } }, ] } } ]Goodtables will now produce these constraints validation errors (Rows 4402 and 4403 only).
$ goodtables download_DP/datapackage.json ... [4402,72] [required-constraint] Column 72 is a required field, but row 4402 has no value [4402,136] [required-constraint] Column 136 is a required field, but row 4402 has no value [4403,135] [maximum-constraint] The value "3536.0" in row 4403 and column 135 does not conform to the maximum constraint of "1000" [4403,136] [required-constraint] Column 136 is a required field, but row 4403 has no value ...